Progress update: Our latest Alfafold model shows a significantly improvement in accuracy and extends the range beyond protein to other biological molecules, including ligand
Since his release in 2020, Alphafold has revolutionized how proteins and their interactions are understood. Google Deepmind and Isomorphic laboratories They worked together to build the foundations of a stronger AI model, which extends the range beyond only protein to the full range of biologically significant molecules.
Today we are sharing updates on progress towards the next generation Alphafold. Our latest model can now generate forecasts of almost all molecules in Protein data bank (PDB), often achieving nuclear accuracy.
It unlocks new understanding and significantly improves accuracy in many key biomolecules classes, including ligands (small molecules), proteins, nucleic acids (DNA and RNA) and those containing post -translational modifications (PTMS). These different types and complexes of structure are necessary to understand the biological mechanisms in the cell and were difficult to predict with high accuracy.
Extended possibilities and performance of the model can help accelerate biomedical breakthroughs and realize the next era of “digital biology” – giving a new insight into the functioning of the disease routes, genomics, biorenevable materials, plant resistance, potential therapeutic goals, drug design mechanisms and new platforms for protein engineering and synthetic biology engineering.
A series of anticipated structures compared to the ground (white) truth from our latest Alphafold model.
Apart from folding protein
Alphafold was a fundamental breakthrough for predicting a single chain protein. Alphafold-Multimer Then it expanded to complexes with many protein chains, and then Alfafold2.3, which improved performance and extended the range to larger complexes.
In 2022, the forecasts of the Alphafold structure for almost all cataloged proteins known to science were freely made available through Alfafold protein structure databasein cooperation with the EMBL's European Bioinformatics Institute (EMBL-EBI).
Until now, 1.4 million users in over 190 countries have gained access to the Alphafold database, and scientists around the world have used Alphafold's forecasts to help develop research on everything, from accelerating new malaria vaccines and the progress of discovering cancer drugs to develop enzymes eating plasticity to solve pollution.
Here, we show the extraordinary Alphafold's abilities to predict accurate structures except for folding protein, generating highly accurate forecasts of structure in ligands, proteins, nucleic acids and coverage modifications.
Performance in protein-league (A), protein (B), nucleic acids (C) and covalent modifications (D).
Acceleration of drug discovery
Early analysis also shows that our model significantly exceeds Alfafold2.3 on some problems of forecasting protein structure that is important for discovering drugs, such as antibodies. In addition, the accurate prediction of the structures of the protein league is an extremely valuable tool for discovering drugs, because it can help scientists identify and design new molecules that can become drugs.
The current industry standard consists in the use of “docking methods” to determine the interaction between ligands and proteins. These docking methods require a rigid structure of the reference protein and a suggested position for the ligand to bind to.
Our latest model sets a new forecasting bar of the protein legal structure by surpassing the best reported methods of docking, without the need for a reference protein structure or location of the ligandu-pocket pocket, and the forecasts of completely new proteins, which were not structurally characterized before.
He can also model the positions of all atoms together, allowing them to represent full -inherent elasticity of proteins and nucleic acids during impact with other molecules – something impossible with the help of docking methods.
Here, for example, there are three recently published, therapeutically important cases in which the expected structures of our latest model (shown in color) closely correspond to the experimentally defined structures (shown in gray):
- Porcno: Anti -cancer molecule of the clinical stage associated with its purpose, along with another protein.
- Kars: Triple complex with covalent ligand (molecular glue) of an important target of cancer.
- Pixed4p4kγ: Selective allosteric lipid kinase inhibitor, with many implications of the disease, including cancer and immune disorders.
Forecasts for Porcn (1), Kras (2) and PI5P4Kγ (3).
Isomorphic laboratories use this new generation alpofold model for therapeutic design of drugs, helping quickly and accurately characterize many types of macromolecular structures important for the treatment of the disease.
New understanding of biology
By unlocking the modeling of protein and ligand structures along with nucleic acids and those containing post -translational modifications, our model is a faster and accurate tool for researching fundamental biology.
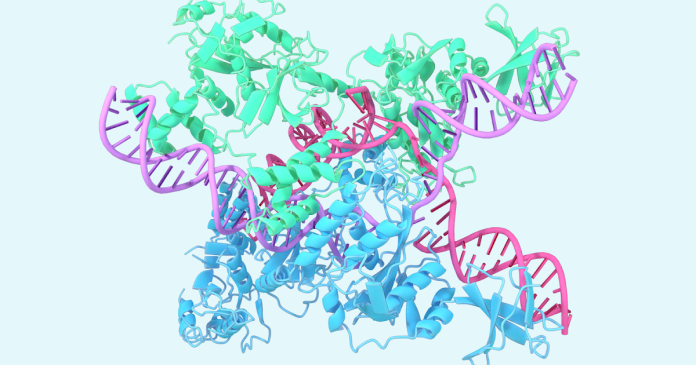
One example includes the structure Caslambda associated with Crrna and DNAHi CRISPR family. Caslambda divides the ability to edit the genome CRISPR-CAS9 SYSTEMCommonly known as “genetic scissors”, which scientists can use to change the bottom of animals, plants and microorganisms. The smaller size of Caslambdy can allow more efficient use in the genome edition.
The expected CaslamBDA (CAS12L) structure associated with CRRNA and DNA, part of the CRISPR subsystem.
The latest version of Alphafold's ability to model such complex systems shows us that AI can help us better understand this type of mechanisms and accelerate their use for therapeutic applications. More examples are Available in our progress update.
Scientific proceedings
The dramatic jump of our model in performance shows AI's potential to significantly increase the scientific understanding of molecular machinery that create a human body – and a wider world of nature.
Alphafold has already catalyzed serious scientific progress around the world. Now the next generation of Alphafold can help in the development of scientific exploration at digital speed.
Our dedicated teams at Google Deepmind and ISOMORFIC Labs have made great progress in this critical work and we are waiting for our further progress to share.